Chapter 1: Introduction to Proteomics—References
Abbott A. 1999. Alliance of US labs plans to build map of cell signalling pathways. Nature 402: 219–220.
Adams J.A. 2001. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101: 2271–2290.
Adams P.D., Seeholzer S., and Ohh M. 2002. Identification of associated proteins by coimmunoprecipitation. In Protein–protein interactions: A molecular cloning manual (ed. E. Golemis), pp. 59–74. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Adamczyk M., Gebler J.C., and Wu J. 2001. Selective analysis of phosphopeptides within a protein mixture by chemical modification, reversible biotinylation and mass spectrometry. Rapid Commun. Mass Spectrom. 15: 1481–1488.
Aebersold R. and Goodlett D.R. 2001. Mass spectrometry in proteomics. Chem. Rev. 101: 269–295.
Ahn N.G. and Resing K.A. 2001. Toward the phosphoproteome. Nat. Biotechnol. 19: 317–318.
Allen N.P., Huang L., Burlingame A., and Rexach M. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276: 29268–29274.
Anderson L. and Seilhamer J. 1997. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18: 533–537.
Annan R.S. and Carr S.A. 1996. Phosphopeptide analysis by matrix-assisted desorption time-of-flight mass spectrometry. Anal. Chem. 68: 3413–3421.
Arenkov P., Kukhtin A., Gemmell A., Voloshchuk S., Chupeeva V., and Mirzabekov A. 2000. Protein microchips: Use for immunoassay and enzymatic reactions. Anal. Biochem. 278: 123–131.
Bader G.D. and Hogue C.W. 2000. BIND—A data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics 16: 465–477.
Baltimore D. 2001. Our genome unveiled. Nature 409: 814–816.
Black D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370.
Blackstock W.P. and Weir M.P. 1999. Proteomics: Quantitative and physical mapping of cellular proteins. Trends Biotechnol. 17: 121–127.
Bode W. and Huber R. 1992. Natural protein proteinase inhibitors and their interactions with proteinases. Eur. J. Biochem. 204: 433–452.
Bowtell D. and Sambrook J. 2003. DNA microarrays: A molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Braun P., Hu Y., Shen B., Halleck A., Koundinya M., Harlow E., and LaBaer J. 2002. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. 99: 2654–2659.
Brenner S.E. 1999a. Errors in genome annotation. Trends Genet. 15: 132–133.
_______. 1999b. Theoretical biology in the third millennium. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354: 1963–1965.
Brody E.N. and Gold L. 2000. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 74: 5–13.
Browne T.R., Van Langenhove A., Costello C.E., Biemann K., and Greenblatt D.J. 1981. Kinetic equivalence of stable-isotope-labeled and unlabeled phenytoin. Clin. Pharmacol. Ther. 29: 511–515.
Burgess A.W., Metcalf D., Sparrow L.G., Simpson R.J., and Nice E.C. 1986. Granulocyte/macrophage colony-stimulating factor from mouse lung conditioned medium: Purification of multiple forms and radioiodination. Biochem. J. 235: 805–814.
Cagney G. and Emili A. 2002. De novo peptide sequencing and quantitative profiling of complex protein mixtures using mass-coded abundance tagging. Nat. Biotechnol. 20: 163–170.
Cahill D.J. 2000. Protein arrays: A high-throughput solution for proteomics research? Proteomics: A Trends Guide 1: 47–51.
Carr S.A., Huddleston M.J., and Annan R.S. 1996. Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Anal. Biochem. 239: 180–192.
Catimel B., Ritter G., Welt S., Old L.J., Cohen L., Nerrie M.A., White S.J., Heath J.K., Demediuk B., Domagala T., Lee F.T., Scott A.M., Tu G.-F., Ji H., Moritz R.L., Simpson R.J., Burgess A.W., and Nice E.C. 1996. Purification and characterization of a novel restricted antigen expressed by normal and transformed human colonic epithelium. J. Biol. Chem. 271: 25664–25670.
Chin E.T. and Papac D.I. 1999. The use of porous graphitic carbon column for desalting hydrophilic peptides prior to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 273: 179–185.
Cole A.R., Ji H., and Simpson R.J. 2000. Proteomic analysis of colonic crypts from normal, multiple intestinal neoplasia and p53-null mice: A comparison with colonic polyps. Electrophoresis 21: 1772–1781.
Conrads T.P., Alving K., Veenstra T.D., Belov M.E., Anderson G.A., Anderson D.J., Lipton M.S., Pasa-Tolic L., Udseth H.R., Chrisler W.B., and Smith R.D. 2001. Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal. Chem. 73: 2132–2139.
Cutler R.L., Metcalf D., Nicola N.A., and Johnson G.R. 1985. Purification of a multipotential colony-stimulating factor from pokeweed mitogen-stimulated mouse spleen cell conditioned medium. J. Biol. Chem. 260: 6579–6587.
Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Otim O., Brown C.T., Livi C.B., Lee P.Y., Revilla R., Rust A.G., Pan Z., Schilstra M.J., Clarke P.J., Arnone M.I., Rowen L., Cameron R.A., McClay D.R., Hood L., and Bolouri H. 2002. A genomic regulatory network for development. Science 295: 1669–1678.
De Leenheer A.P. and Thienpont L.M. 1992. Applications of isotope-dilution mass-spectrometry in clinical-chemistry, pharmacokinetics, and toxicology. Mass Spectrom. Rev. 11: 249–307.
Duan X.J., Xenarios I., and Eisenberg D. 2002. Describing biological protein interactions in terms of protein states and state transitions: The LiveDIP Database. Mol. Cell. Proteomics 1: 104–116.
Dunham I., Shimizu N., Roe B.A., Chissoe S., Dunham I., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., et al. 1999. The DNA sequence of human chromosome 22. Nature 402: 489–495.
Einarson M.B. and Orlinick J.R. 2002. Identification of protein–protein interactions with glutathione-S-transferase fusion proteins. In Protein–protein interactions: A molecular cloning manual (ed. E. Golemis), pp. 37–57. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Eisenberg D., Marcotte E.M., Xenarios I., and Yeates T.O. 2000. Protein function in the post-genomic era. Nature 405: 823–826.
Emmert-Buck M.R., Bonner R.F., Smith P.D., Chuqui R.F., Zhuang Z., Goldstein S.R., Weiss R.A., and Liotta L.A. 1996. Laser capture microdissection. Science 274: 998–1001.
Ficarro S.B., McCleland M.L., Stukenberg P.T., Burke D.J., Ross M.M., Shabanowitz J., Hunt D.F., and White F.M. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20: 301–305.
Frolik C.A., Dart L.L, Meyers C.A., Smith D.M., and Sporn M.B. 1983. Purification and initial characterization of a type 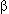 transforming growth factor from human placenta. Proc. Natl. Acad. Sci. 80: 3676–3680. transforming growth factor from human placenta. Proc. Natl. Acad. Sci. 80: 3676–3680.
Gao C., Mao S., Lo C.H., Wirsching P., Lerner R.A., and Janda K.D. 1999. Making artificial antibodies: A format for phage display of combinatorial heterodimeric arrays. Proc. Natl. Acad. Sci. 96: 6025–6030.
Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147.
Gerstein M., Lan N., and Jansen R. 2002. Proteomics. Integrating interactomes. Science 295: 284–287.
Goldman R.D. 2000. Antibodies: Indispensable tools for biomedicial research. Trends Biochem. Sci. 25: 593–595.
Goodlett D.R., Keller A., Watts J.D., Newitt R., Yi E.C., Purvine S., Eng J.K., von Haller P., Aebersold R., and Kolker E. 2001. Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun. Mass Spectrom. 15: 1214–1221.
Görg A., Postel W., Domscheit A., and Gunther S. 1988. Two-dimensional electrophoresis with immobilized pH gradients of leaf proteins from barley (Hordeum vulgare): Method, reproducibility and genetic aspects. Electrophoresis 9: 681–692.
Görg A., Obermaier C., Boguth G., Harder A., Scheibe B., Wildgruber R., and Weiss W. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21: 1037–1053.
Goshe M.B., Conrads T.P., Panisko E.A., Angell N.H., Veenstra T.D., and Smith R.D. 2001. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Anal. Chem. 73: 2578–2586.
Gospodarowicz D., Cheng J., Lui G.-M., Baird A., and Bohlent P. 1984. Isolation of brain fibroblast growth factor by heparin-Sepharose affinity chromatography: Identity with pituitary fibroblast growth factor. Proc. Natl. Acad. Sci. 81: 6963–6967.
Graeber T.G. and Eisenberg D. 2001. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat. Genet. 29: 295–300.
Graziano M.P., Moxham C.P., and Malbon C.C. 1985. Purified rat hepatic 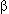 2-adrenergic receptor. J. Biol. Chem. 260: 7665–7674. 2-adrenergic receptor. J. Biol. Chem. 260: 7665–7674.
Green L.S., Bell C., and Janjic N. 2001. Aptamers as reagents for high-throughput screening. BioTechniques 30: 1094–1096.
Greis K.D., Hayes B.K., Comer F.I., Kirk M., Barnes S., Lowary T.L., and Hart G.W. 1996. Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry. Anal. Biochem. 234: 38–49.
Griffin T.J., Goodlett D.R., and Aebersold R. 2001. Advances in proteome analysis by mass spectrometry. Curr. Opin. Biotechnol. 12: 607–612.
Gunneriussion E., Nord K., Uhlen M., and Nygren P. 1999. Affinity maturation of a Taq DNA polymerase specific affibody by helix shuffling. Protein Eng. 12: 873–878.
Gunneriusson E., Samuelson P., Ringdahl J., Gronlund H., Nygren P.A., and Stahl S. 1999. Staphylococcal surface display of immunoglobulin A (IgA)- and IgE-specific in vitro-selected binding proteins (affibodies) based on Staphylococcus aureus protein A. Appl. Environ. Microbiol. 65: 4134–4140.
Gygi S.P., Rochon Y., Franza B.R., and Aebersold R. 1999a. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720–1730.
Gygi S.P., Corthals G.L., Zhang Y., Rochon Y., and Aebersold R. 2000. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. 97: 9390–9395.
Gygi S.P., Han D.K., Gingras A.C., Sonenberg N., and Aebersold R. 1999b. Protein analysis by mass spectrometry and sequence database searching: Tools for cancer research in the post-genomic era. Electrophoresis 20: 310–319.
Gygi S.P., Rist B., Gerber S.A., Turecek F., Gelb M.H., and Aebersold R. 1999c. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17: 994–999.
Haga K. and Haga T. 1985. Purification of the muscarinic acetylcholine receptor from porcine brain. J. Biol. Chem. 260: 7927–7935.
Hatzimanikatis V. and Lee K.H. 1999. Dynamical analysis of gene networks requires both mRNA and protein expression information. Metab. Eng. 1: 275–281.
Hazbun T.R. and Fields S. 2001. Networking proteins in yeast. Proc. Natl. Acad. Sci. 98: 4277–4278.
Heinemeyer T., Chen X., Karas H., Kel A.E., Kel O.V., Liebich I., Meinhardt T., Reuter I., Schacherer F., and Wingender E. 1999. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27: 318–322.
Heldin C.-H., Westermark B., and Wasteson A. 1981. Platelet-derived growth factor. Biochem. J. 193: 907–913.
Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A.R., Sassi H., Nielsen P.A., Rasmussen K.J., Andersen J.R., Johansen L.E., Hansen L.H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sorensen B.D., Matthiesen J., Hendrickson R.C., Gleeson F., Pawson T., Moran M.F., Durocher D., Mann M., Hogue C.W., Figeys D., and Tyers M. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183.
Hoogland C., Sanchez J.C., Walther D., Baujard V., Baujard O., Tonella L., Hochstrasser D.F., and Appel R.D. 1999. Two-dimensional electrophoresis resources available from ExPASy. Electrophoresis 20: 3568–3571.
Hoppe-Seyler F. and Butz K. 2000. Peptide aptamers: Powerful new tools for molecular medicine. J. Mol. Med. 78: 426–430.
Houry W.A., Frishman D., Eckerskorn C., Lottspeich F., and Hartl F.U. 1999. Identification of in vivo substrates of the chaperonin GroEL. Nature 402: 147–154.
Huddleston M.J., Annan R.S., and Carr S.A. 1993. Selective detection of phosphopeptides in complex mixtures by electrospray liquid chromatography. J. Am. Soc. Mass Spectrom. 4: 710–717.
Hunter A.P. and Games D.E. 1994. Chromatographic and mass spectrometric methods for the identification of phosphorylation sites in phosphoproteins. Rapid Commun. Mass Spectrom. 8: 559–570.
Hunter T. 2000a. The role of tyrosine phosphorylation in cell growth and disease. Harvey Lect. 94: 81–119.
_______. 2000b. Signaling—2000 and beyond. Cell 100: 113–127.
Huse M. and Kuriyan J. 2002. The conformational plasticity of protein kinases. Cell 109: 275–282.
Ideker T., Thorsson V., Ranish J.A., Christmas R., Buhler J., Eng J.K., Bumgarner R., Goodlett D.R., Aebersold R., and Hood L. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934.
Ito T., Chiba T., and Yoshida M. 2001a. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol. (suppl.) 19: S23–S27.
Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., and Sakaki Y. 2001b. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569–4574.
Jayasena S.D. 1999. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45: 1628–1650.
Johnson L.N. and Lewis R.J. 2001. Structural basis for control by phosphorylation. Chem. Rev. 101: 2209–2242.
Kanchisa M. and Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30.
Karp P.D., Riley M., Saier M., Paulsen I.T., Paley S., Pellegrini-Toole A., Bonavides C., and Gama-Castro S. 2002. The EcoCyc Database. Nucleic Acids Res. 30: 56–58.
Kemp B.E. and Pearson R.B. 1991. Intrasteric regulation of protein kinases and phosphatases. Biochim. Biophys. Acta 1094: 67–76.
Khan A.R. and James M.N. 1998. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 7: 815–836.
Kim S.H. 2000. Structural genomics of microbes: An objective. Curr. Opin. Struct. Biol. 10: 380–383.
Kitano H. 2002. Systems biology: A brief overview. Science 295: 1662–1664.
Klose J. 1975. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik. 26: 231–243.
Knappik A., Ge L., Honegger A., Pack P., Fischer M., Wellnhofer G., Hoess A., Wolle J., Pluckthun A., and Virnekas B. 2000. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular concensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 296: 57–86.
Kobe B. and Kemp B.E. 1999. Active site-directed protein regulation. Nature 402: 373–376.
Krebs B., Rauchenberger R., Reiffert S., Rothe C., Tesar M., Thomassen E., Cao M., Dreier T., Fischer D., Hoss A., et al. 2001. High-throughput generation and engineering of recombinant human antibodies. J. Immunol. Methods 254: 67–84.
Kreider B.L. 2000. PROfusion: Genetically tagged proteins for functional proteomics and beyond. Med. Res. Rev. 20: 212–215.
Krishna R. and Wold F. 1997. Identification of common post-translational modifications. In Protein structure—A practical approach, 2nd edition (ed. T.E. Creighton), pp. 91–116. Oxford University Press, New York.
Kussmann M., Hauser K., Kissmehl R., Breed J., Plattner H., and Roepstorff P. 1999. Comparison of in vivo and in vitro phosphorylation of the exocytosis-sensitive protein PP63/parafusin by differential MALDI mass spectrometric peptide mapping. Biochemistry 38: 7780–7790.
Kuster B. and Mann M. 1999. 18O-labeling of N-glycosylation sites to improve the identification of gel-separated glycoproteins using peptide mass mapping and database searching. Anal. Chem. 71: 1431–1440.
Lakey J.H. and Raggett E.M. 1998. Measuring protein-protein interactions. Curr. Opin. Struct. Biol. 8: 119–123.
Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921.
Lahm H.W. and Langen H. 2000. Mass Spectrometry: A tool for the identification of proteins separated by gels. Electrophoresis 21: 2105–2114.
Larsen M.R., Sorensen G.L., Fey S.J., Larsen P.M., and Roepstorff P. 2001. Phospho-proteomics: Evaluation of the use of enzymatic de-phosphorylation and differential mass spectrometric peptide mass mapping for site specific phosphorylation assignment in proteins separated by gel electrophoresis. Proteomics 1: 223–238.
Lecerf J.M., Shirley T.L., Zhu Q., Kazantsev A., Amersdorfer P., Housman D.E., Messer D.E., and Huston J.S. 2001. Human single-chain Fv intrabodies counteract in situ huntingtin aggregation in cellular models of Huntington's disease. Proc. Natl. Acad. Sci. 98: 4764–4769.
Lee M. and Walt D.R. 2000. A fiber-optic microarray biosensor using aptamers as receptors. Anal. Biochem. 282: 142–146.
Legrain P. 2002. Protein domain networking. Nat. Biotechnol. 20: 128–129.
Legrain P. and Selig L. 2000. Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett. 480: 32–36.
Legrain P., Wojcik J., and Gauthier J.M. 2001. Protein–protein interaction maps: A lead towards cellular functions. Trends Genet. 17: 346–352.
Li M. 2000. Applications of display technology in protein analysis. Nat. Biotechnol. 18: 1251–1256.
Liao P.C., Leykam J., Andrew P.C., Gage D.A., and Allison J. 1994. An approach to locate phosphorylation sites in a phosphoprotein: Mass mapping by combining specific enzymatic degradation with matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 219: 9–20.
Link A.J., Eng J., Schieltz D.M., Carmack E., Mize G.J., Morris D.R., Garvik B.M., and Yates J.R. III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17: 676–682.
Lockhart D.J. and Winzeler E.A. 2000. Genomics, gene expression and DNA arrays. Nature 405: 827–836.
Lohse P.A. and Wright M.C. 2001. In vitro protein display in drug discovery. Curr. Opin. Drug Disc. Dev. 4: 198–204.
MacBeath G. and Schreiber S.L. 2000. Printing proteins as microarrays for high-throughput function determination. Science 289: 1760–1763.
MacBeath G., Koehler A.N., and Schreiber S.L. 1999. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J. Am. Chem Soc. 121: 7967–7968.
Mann M., Ong S.E., Gronborg M., Steen H., Jensen O.N., and Pandey A. 2002. Analysis of protein phosphorylation using mass spectrometry: Deciphering the phosphoproteome. Trends Biotechnol. 20: 261–268.
Matsumoto H., Kahn E.S., and Komori N. 1997. Separation of phosphopeptides from their non-phosphorylated forms by reversed-phase POROS perfusion chromatography at alkaline pH. Anal. Biochem. 251: 116–119.
Mayer M.L. and Hieter P. 2000. Protein networks—Built by association. Nat. Biotechnol. 18: 1242–1243.
McLafferty F.W., Fridriksson E.K., Horn D.M., Lewis M.A., and Zubarev R.A. 1999. Techview: Biochemistry. Biomolecule mass spectrometry. Science 284: 1289–1290.
Mellor J.C., Yanai I., Clodfelter K.H., Mintseris J., and DeLisi C. 2002. Predictome: A database of putative functional links between proteins. Nucleic Acids Res. 30: 306–309.
Mewes H.W., Frishman D., Guldener U., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., and Weil B. 2002. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 30: 31–34.
Meyer H.E., Eisermann B., Heber M., Hoffmann-Posorke E., Korte H., Weigt C., Wegner A., Hutton T., Donella-Deana A., and Perich J.W. 1993. Strategies for nonradioactive methods in the localization of phosphorylated amino acids in proteins. FASEB J. 7: 776–782.
Mirgorodskaya O.A., Kozmin Y.P., Titov M.L., Korner R., Sonsken C.P., and Roepstorff P. 2000. Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun. Mass Spectrom. 14: 1226–1232.
Mironov A.A., Fickett J.W., and Gelfand M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288–1293.
Monod J., Changeux J.P., and Jacob F. 1963. Allosteric proteins and cellular control systems. J. Mol. Biol. 6: 306–329.
Munchbach M., Quadroni M., Miotto G., and James P. 2000. Quantitation and facilitated de novo sequencing of proteins by isotopic N-terminal labeling of peptides with a fragmentation-directing moiety. Anal. Chem. 72: 4047–4057
Nature 1999. Proteomics, transcriptomics: What's in a name? Nature 402: 715.
Neubauer G. and Mann M. 1999. Mapping of phosphorylation sites of gel-isolated proteins by nanoelectrospray tandem mass spectrometry: Potentials and limitations. Anal. Chem. 71: 235–242.
Neubauer G., Gottschalk A., Fabrizio P., Séraphin B., Luhrmann R., and Mann M. 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. 94: 385–390.
Neubauer G., King A., Rappsilber J., Calvio C., Watson M., Ajuh P., Sleeman J., Lamond A., and Mann M. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20: 46–50.
Nicola A., Metcalf D., Matsumoto M., and Johnson G.R. 1983. Purification of a factor inducing differentiation in murine myelomonocytic leukemia cells—Identification as granulocyte colony stimulating factor. J. Biol. Chem. 258: 9017–9023.
Oda Y., Nagasu T., and Chait B.T. 2001. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 19: 379–382.
Oda Y., Huang K., Cross F.R., Cowburn D., and Chait B.T. 1999. Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. 96: 6591–6596.
O'Farrell P.H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250: 4007–4021.
Oliver S.G. 2002. Functional genomics: Lessons from yeast. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 17–23.
Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., and Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1: 376–386.
Patterson S.D. 2000a. Proteomics: The industrialization of protein chemistry. Curr. Opin. Biotechnol. 11: 413–418.
_______. 2000b. Mass spectrometry and proteomics. Physiol. Genomics 2: 59–65.
Patton W.F. 2000. A thousand points of light: The application of fluorescence detection technologies to two-dimensional gel electrophoresis and proteomics. Electrophoresis 21: 1123–1144.
Pawson T. and Nash P. 2000. Protein-protein interactions define specificity in signal transduction. Genes Dev. 14: 1027–1047.
Pelletier J. and Sidhu S. 2001. Mapping protein-protein interactions with combinatorial biology methods. Curr. Opin. Biotechnol. 12: 340–347.
Porath J. 1992. Immobilized metal ion affinity chromatography. Protein Expr. Purif. 3: 263–281.
Posewitz M.C. and Tempst P. 1999. Immobilized gallium (III) affinity chromatography of phosphopeptides. Anal. Chem. 71: 2883–2892.
Pugh B.F. 1996. Mechanisms of transcription complex assembly. Curr. Opin. Cell. Biol. 8: 303–311.
Raamsdonk L.M., Teusink B., Broadhurst D., Zhang N., Hayes A., Walsh M.C., Berden J.A., Brindle K.M., Kell D.B., Rowland J.J., Westerhoff H.V., van Dam K., and Oliver S.G. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19: 45–50.
Raum T., Gruber R., Riethmuller G., and Kufer P. 2001. Anti-self antibodies selected from a human IgD heavy chain repertoire: A novel approach to generate therapeutic human antibodies against tumor-associated differentiation antigens. Cancer Immunol. Immunother. 50: 141–150.
Ray L.B. and Gough N.R. 2002. Orienteering strategies for a signaling maze. Science 296: 1632–1633.
Regnier F.E., Riggs L., Zhang R., Xiong L., Liu P., Chakraborty A., Seeley E., Stoma C., and Thompson R.A. 2002. Comparative proteomics based on stable isotope labeling and affinity selection. J. Mass Spectrom. 37: 133–145.
Resing K.A. and Ahn N.G. 1997. Protein phosphorylation analysis by electrospray ionization-mass spectrometry. Methods Enzymol. 283: 29–44.
Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., and Chait B.T. 2000. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651.
Rubinstein M., Rubinstein S., Familetti P.C., Miller R.S., Waldman A.A., and Pestka S. 1979. Human leukocyte interferon: Production, purification to homogeneity, and initial characterization. Proc. Natl. Acad. Sci. 76: 640–644.
Ryu D.D. and Nam D.H. 2000. Recent progress in biomolecular engineering. Biotechnol. Prog. 16: 2–16.
Schaller H.C. and Bodenmuller H. 1981. Isolation and amino acid sequence of a morphogenetic peptide from hydra. Proc. Natl. Acad. Sci. 78: 7000–7004.
Schwikowski B., Uetz P., and Fields S. 2000. A network of protein-protein interactions in yeast. Nat. Biotechnol. 18: 1257–1261.
Séraphin B., Puig O., Bouveret E., Rutz B., and Caspary F. 2002. Tandem affinity purification to enhance interacting protein identification. In Protein–protein interactions: A molecular cloning manual (ed. E. Golemis), pp. 313–328. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Shi Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9: 459–470.
Sickmann A. and Meyer H.E. 2001. Phosphoamino acid analysis. Proteomics 1: 200–206.
Simpson R.J. and Dorow D.S. 2001. Cancer proteomics: From signaling networks to tumor markers. Trends Biotechnol. 19: S40–S48.
Simpson R.J. and Nice E.C. 1989. Strategies for the purification of subnanomole amounts of protein and polypeptides for microsequence analysis. In The use of HPLC in receptor biochemistry, pp. 201–244. A.R Liss, New York.
Stensballe A., Andersen S., and Jensen O.N. 2000. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics 1: 207–222.
Strohman R. 1994. Epigenesis: The missing beat in biotechnology? Bio/Technology 12: 156–164.
Sze S.K., Ge Y., Oh H., and McLafferty F.W. 2002. Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc. Natl. Acad. Sci. 99: 1774–1779.
Tatemoto K. 1982. Isolation and characterization of peptide YY (PYY), a candidate gut hormone that inhibits pancreatic exocrine secretion. Proc. Natl. Acad. Sci. 79: 2514–2518.
Templin M.F., Stoll D., Schrenk M., Traub P.C., Vohringer C.F., and Joos T.O. 2002. Protein microarray technology. Trends Biotechnol. 20: 160–166.
Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., and Rothberg J.M. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627.
Unlu M., Morgan M.E., and Minden J.S. 1997. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis 18: 2071–2077.
van Driel I.R., Stearn P.A., Grego B., Simpson R.J., and Goding J.W. 1984. The receptor for transferrin on murine myeloma cells one step purification based on its physiology and partial amino acid sequence. J. Immunol. 133: 3220–3224.
van Helden J., Naim A., Mancuso R., Eldridge M., Wernisch L., Gilbert D., and Wodak S.J. 2000. Representing and analysing molecular and cellular function using the computer. Biol. Chem. 381: 921–935.
Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A, et al. 2001. The sequence of the human genome. Science 291: 1304–1351.
Verhagen A.M., Ekert P.G., Pakusch M., Silke J., Connolly L.M., Reid G.E., Moritz R.L., Simpson R.J., and Vaux D.L. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53.
Verma R., Annan R.S., Huddleston M.J., Carr S., Reynard G., and Deshaies R.J. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460.
Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., and Deshaies R.J. 2000. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11: 3425–3439.
Wang A.M. and Creasey A.A. 1985. Molecular cloning of the complementary DNA for human tumor necrosis factor. Science 228: 149–154.
Wells L., Vosseller K., and Hart G.W. 2001. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 291: 2376–2378.
Weng G., Bhalla U.S., and Iyengar R. 1999. Complexity in biological signaling systems. Science 284: 92–96.
Wilson D.S., Keefe A.D., and Szostak J.W. 2001. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. 98: 3750–3755.
Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Prüss M., Reuter I., and Schacherer F. 2000. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 28: 316–319.
Wool I.G., Chan Y.L., and Gluck A. 1995. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 73: 933–947.
Xenarios L. and Eisenberg D. 2001. Protein interaction databases. Curr. Opin. Biotechnol. 12: 334–339.
Xenarios I., Salwínski L., Duan X.J., Higney P., Kim S.-M., and Eisenberg D. 2002. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30: 303–305.
Xhou W., Merrick B.A., Khaledi M.G., and Tomer K.B. 2000. Detection and sequencing of phosphopeptides affinity bound to immobilized metal ion beads by matrix-assisted laser desorption mass spectrometry. J. Am. Soc. Mass Spectrom. 11: 273–282.
Yan J.X., Packer N.H., Gooley A.A., and Williams K.L. 1998. Protein phosphorylation: Technologies for the identification of phosphoamino acids. J. Chromatogr. A 808: 23–41.
Yao X., Freas A., Ramirez J., Demirev P.A., and Fenselau C. 2001. Proteolytic 18O labeling for comparative proteomics: Model studies with two serotypes of adenovirus. Anal. Chem. 73: 2836–2842.
Yates J.R., III. 1998a. Database searching using mass spectrometry data. Electrophoresis 19: 893–900.
_______. 1998b. Mass spectrometry and the age of the proteome. J. Mass Spectrom. 33: 1–19.
Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., and Cesareni G. 2002. MINT: A Molecular INTeraction database. FEBS Lett. 513: 135–140.
Zhang J.-G., Farley A., Nicholson S.E., Willson T.A., Zugaro L.M., Simpson R.J., Moritz R.L., Cary D., Richardson R., Hausmann G., Kile B.J., Kent S.B.H., Alexander W.S., Metcalf D., Hilton D.J., Nicola N.A., and Baca M. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. 96: 2071–2076.
Zhang X., Herring C.J., Romano P.R., Szczepanowska J., Brzeska H., Hinnebusch A.G., and Qin J. 1998. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal Chem. 70: 2050–2059.
Zhou H., Watts J.D., and Aebersold R. 2001. A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 19: 375–378.
Zhou W., Merrick B.A., Khaledi M.G., and Tomer K.B. 2000. Detection and sequencing of phosphopeptides affinity bound to immobilized metal ion beads by matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectr. 11: 273–282.
Zhou H., Ranish J.A., Watts J.D., and Aebersold R. 2002a. Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nat. Biotechnol. 20: 512–515.
Zhou G., Li H., DeCamp D., Chen S., Shu H., Gong Y., Flaig M., Gillespie J.W., Hu N., Taylor P.R., et al. 2002b. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol. Cell. Proteomics 1: 117–124.
Zhu H., Klemic J.F., Chang S., Bertone P., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., and Snyder M. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26: 283–289.
Zugaro L.M., Reid G.E., Ji H., Eddes J.S., Murphy A.C., Burgess A.W., and Simpson R.J. 1998. Characterization of rat brain stathmin isoforms by two-dimensional gel electrophoresis-matrix assisted laser desorption/ionization and electrospray ionization-ion trap mass spectrometry. Electrophoresis 19: 867–876.
|

