Chapter 10: Characterization of Protein Complexes—References
Aboul-Enein A. and Schulte-Frohlinde D. 1988. Biological deactivation and single-strand breakage of plasmid DNA by photosensitization using tris(2,2'-bipyridyl)ruthenium(II) and peroxydisulfate. Photochem. Photobiol. 48: 27–34.
Allen N.P., Huang L., Burlingame A., and Rexach M. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276: 29268–29274.
Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., and Lamond A.I. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12: 1–11.
Aravind L. 2000. Guilt by association: Contextual information in genome analysis. Genome Res. 10: 1074–1077.
Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., and Struhl K., eds. 1997. Current protocols in molecular biology. Wiley, New York.
Bader G.D., Donaldson I., Wolting C., Ouellette B.F., Pawson T., and Hogue C.W. 2001. BIND—The Biomolecular Interaction Network Database. Nucleic Acids Res. 29: 252–245.
Baldwin R.L. 1996. How Hofmeister ion interactions affect protein stability. Biophys. J. 71: 2056–2063.
Bartel P.L., Roecklein J.A., SenGupta D., and Fields S.A. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12: 72–77.
Bartholomew B., Kassavetis G., Braun B., and Geiduschek E.P. 1990. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photo-cross-linking reagent. EMBO J. 9: 2197–2205.
Bartlett M.S., Thomm M., and Geiduschek E.P. 2000. The orientation of DNA in an archaeal transcription initiation complex. Nat. Struct. Biol. 7: 782–785.
Bell S. and Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134.
Black D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370.
Blom J., Dekker P.J.T., and Meijer M. 1995. Functional and physical interactions of components of the yeast mitochondrial inner-membrane import machinery (MIM). Eur. J. Biochem. 232: 309–314.
Bolletta F., Juris A., Maestri M., and Sandrini D. 1980. Quantum yield of formation of the lowest excited state of Ru(bpy)2+3 and Ru(phen)2+3. Inorg. Chim. Acta 44: L175–L176.
Boussif O., Lezoualc'h F., Zanta M.A, Mergny M.D., Scherman D., Demeneix B., and Behr J.-P. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. 92: 7297–7301.
Cantor C.R. and Schimmel P.R. 1980. Biophysical chemistry, part III. The behavior of biological molecules. W.H. Freeman, San Francisco, California.
Carey J. 1991. Gel retardation. Methods Enzymol. 208: 103–117.
Chen Z. and Han M. 2000. Building a protein interaction map: Research in the post-genome era. BioEssays 22: 503–506.
Clegg R.M. 1996. Fluorescence resonance energy transfer. In Fluorescence imaging spectroscopy and microscopy (ed. X.F. Wang and B. Herman), pp. 179–252. Wiley, New York.
Connelly P.R., Varadarajan R., Sturtevant J.M., and Richards F.M. 1990. Thermodynamics of protein–peptide interactions in the ribonuclease S system studied by titration calorimetry. Biochemistry 29: 6108–6114.
Crameri A., Whitehorn E.A., Tate E., and Stemmer W.P.C. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14: 315–319.
Croft L., Schandorff S., Clark F., Burrage K., Arctander P., and Mattick J.S. 2000. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 24: 340–341.
Cubitt A.B., Heim R., Adams S.R., Boyd A.E., Gross L.A., and Tsien R.Y. 1995. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20: 448–455.
Dabbeni-Sala F., Di Santo S., Franceschini D., Skaper S.D., and Giusti P. 2001. Melatonin protects against neurotoxicity in rats: A role for mitochondrial complex I activity. FASEB J. 15: 164–170.
Damelin M. and Silver P.A. 2000. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell 5: 133–140.
_______. 2002. In situ analysis of spatial relationships between proteins of the nuclear pore complex. Biophys. J. (in press).
Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., et al. 2002. A genomic regulatory network for development. Science 295: 1669–1678.
Dekker P.J.T., Ryan M.T., Brix J., Müller H., Hönlinger A., and Pfanner N. 1998. The preprotein translocase of the outer mitochondrial membrane: Molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18: 6515–6524.
Delagrave S., Hawtin R.E., Silva C.M., Yang M.M., and Youvan D.C. 1995. Red-shifted excitation mutants of the green fluorescent protein. Bio/Technology 13: 151–154.
Duan J.D., Xenarios I., and Eisenberg D. 2002. Describing biological protein interactions in terms of protein states and state transitions. Mol. Cell Proteomics 1: 104–116.
Ehrig T., O'Kane D.J., and Prendergast F.G. 1995. Green-fluorescent protein with altered fluorescence excitation spectra. FEBS Lett. 367: 163–166.
Ellis R.J. 2001. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 26: 597–604.
Fancy D.A. 2000. Elucidation of protein–protein interactions using chemical cross-linking or label transfer techniques. Curr. Opin. Chem.Biol. 4: 28–33.
Fancy D.A. and Kodadek T. 1999. New chemistry for the analysis of protein-protein interactions: Rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. 96: 6020–6024.
Fancy D.A., Denison C., Kim K., Xie Y., Holdeman T., Amini F., and Kodadek T. 2000. Scope, limitations and mechanistic aspects of the photo-induced cross-linking of proteins by water-soluble metal complexes. Chem. Biol. 7: 697–708.
Fidanza J., Ozaki H., and McLaughlin L. 1992. Site-specific labeling of DNA sequences containing phosphorothioate diesters. J. Amer. Chem. Soc. 114: 5509–5517.
Fields S. and Song O.K. 1989. A novel genetic system to detect protein–protein interactions. Nature 340: 245–246.
Fisher B.M., Ha J.-H., and Raines R.T. 1998. Coulombic forces in protein-RNA interactions: Binding and cleavage by ribonuclease A and variants at Lys7, Arg10 and Lys66. Biochemistry 37: 12121–12132.
Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspe G., Tiollais P., Transy C., and Legrain P. 2000. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242: 369–379.
Flores A., Briand J.F., Gadal O., Andrau J.C., Rubbi L., Van Mullem V., Boschiero C., Goussot M., Marck C., Carles C., et al. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. 96: 7815–7820.
Forwood J.K., Lam M.H.C., and Jans D.A. 2001. Nuclear import of Creb and AP-1 transcription factors requires importin-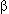 1 and Ran but is independent of importin- 1 and Ran but is independent of importin- . Biochemistry 40: 5208–5217. . Biochemistry 40: 5208–5217.
Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., and Lamond A.I. 2002. Paraspeckles: A novel nuclear domain. Curr. Biol. 12: 13–25.
Franken K.L., Hiemstra H.S., van Meijgaarden K.E., Subronto Y., den Hartigh J., Ottenhoff T.H., and Drijfhout J.W. 2000. Purification of his-tagged proteins by immobilized chelate affinity chromatography: The benefits from the use of organic solvent. Protein Expr. Purif. 18: 95–99.
Garin J., Diez R., Kieffer S., Dermine J.F., Duclos S., Gagnon E., Sadoul R., Rondeau C., and Desjardins M. 2001. The phagosome proteome: Insight into phagosome functions. J. Cell Biol. 152: 165–180.
Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M.A., Copley R.R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., and Superti-Furga G. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147.
Gerstein M., Lan N., and Jansen R. 2002. Integrating interactomes. Science 295: 284–285.
Gordon G.W., Berry G., Liang X.H., Levine B., and Herman B. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74: 2702–2713.
Guthrie C. and Fink G. 2002. Guide to yeast genetics and molecular cell biology, parts B, C. Methods Enzymol., vols. 350, 351. Academic Press, San Diego, California.
Hames B.D. 1990. One-dimensional polyacrylamide gel electrophoresis. In Gel electrophoresis of proteins: A practical approach, 2nd edition (ed. B.D. Hames and D. Rickwood), pp. 1–148. IRL Press/Oxford University Press, Oxford, United Kingdom.
Han D.K., Eng J., Zhou H., and Aebersold R. 2001. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19: 946–951.
Hanes J. and Pluckthun A. 1997. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. 94: 4937–4042.
Heim R. and Tsien R.Y. 1996. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6: 178–182.
Heim R., Cubitt A.B., and Tsien R.Y. 1995. Improved green fluorescence. Nature 373: 663–664.
Hermanson G.T. 1996. Bioconjugate techniques. Academic Press.
Heyduk T., Ma Y., Tang H., and Ebright R.H. 1996. Fluorescence anisotropy: Rapid, quantitative assay for protein–DNA and protein–protein interaction. Methods Enzymol. 274: 492–503.
Hilton D.J., Richardson R.T., Alexander W.S., Viney E.M., Willson T.A., Sprigg N.S., Starr R., Nicholson S.E., Metcalf D., and Nicola N.A. 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. 95: 114–119.
Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A.R., Sassi H., Nielsen P.A., Rasmussen K.J., Andersen J.R., Johansen L.E., Hansen L.H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sorensen B.D., Matthiesen J., Hendrickson R.C., Gleeson F., Pawson T., Moran M.F., Durocher D., Mann M., Hogue C.W., Figeys D., and Tyers M. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183.
Hogenesch J.B., Ching K.A., Batalov S., Su A.I., Walker J.R., Zhou Y., Kay S.A., Schultz P.G., and Cooke M.P. 2001. A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell 106: 413–415.
Holzinger A., Phillips K.S., and Weaver T.E. 1996. Single-step purification/solubilization of recombinant proteins: Application to surfactant protein B. BioTechniques 20: 804–808.
Houry W.A., Frishman D., Eckerskorn C., Lottspeich F., and Hartl F.U. 1999. Identification of in vivo substrates of the chaperonin GroEL. Nature 402: 147–154.
Ideker T., Thorsson V., Ranish J.A., Christmas R., Buhler J., Eng J.K., Bumgarner R., Goodlett D.R., Aebersold R., and Hood L. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934.
Ito T., Tashiro K., Muta S., Ozawa R., Chiba T., Nishizawa M., Yamamoto K., Kuhara S., and Sakaki Y. 2000. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. 97: 1143–1147.
Jameson D.M. and Sawyer W.H. 1995. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 246: 283–300.
Kamp R.M. 1988. Application of bifunctional reagents for topological investigation. In Modern methods in protein chemistry (ed. H. Tschesche), vol. 3. pp. 275–298. Walter de Gruyter & Co., Berlin, Germany.
Kiessig S., Reissmann J., Rascher C., Kullertz G., Fischer A., and Thunecke F. 2001. Application of a green fluorescent fusion protein to study protein–protein interactions by electrophoretic methods. Electrophoresis 22: 1428–1435.
Kim K., Fancy D.A., and Kodadek T. 1999. Photo-induced protein cross-linking mediated by palladium porphyrins. J. Am. Chem. Soc. 121: 11896–11897.
Kim R., Yokota H., and Kim S.-H. 1999. Electrophoresis of proteins and protein–protein complexes in a native agarose gel. Anal. Biochem. 282: 147–149.
Kim T.-K., Reinberg D., and Ebright R. 2000a. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288: 1418–1422.
Kim T.-K., Lagrange T., Reinberg D., Naryshkin N., and Ebright R. 2000b. Site-specific protein-DNA photo-cross-linking. In DNA-protein interactions (ed. A. Travers and M. Buckle), pp. 319–335. Oxford University Press, Oxford, United Kingdom.
Kim T.-K., Lagrange T., Wang Y.-H., Griffith J., Reinberg D., and Ebright R. 1997. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc. Natl. Acad. Sci. 94: 12268–12273.
Kitano H. 2002. Systems biology: A brief overview. Science 295: 1662–1664.
Kodadek T., Gan D.C., and Stemke-Hale K. 1989. The phage T4 uvs recombination protein stabilizes presynaptic filaments. J. Biol. Chem. 264: 16451–16457.
Kratchmarova I., Kalume D.E., Blagoev B., Scherer P.E., Podtelejnikov A.V., Molina H., Bickel P.E., Andersen J.S., Fernandez M.M., Bunkenborg J., Roepstorff P., Kristiansen K., Lodish H.F., Mann M., and Pandey A. 2002. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol. Cell. Proteomics 1: 213–222.
Krowcyznska A.M., Donoghue K., and Hughes L. 1995. Recovery of DNA, RNA and protein from gels with microconcentrators. BioTechniques 18: 698–703.
Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
Lagrange T., Kapanidis A., Tang H., Reinberg D., and Ebright R. 1998. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12: 34–44.
Lagrange T., Kim T.K., Orphanides G., Ebright Y., Ebright R., and Reinberg D. 1996. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photo-cross-linking: Organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. 93: 10620–10625.
Lakey J.H. and Raggett E.M. 1998. Measuring protein-protein interactions. Curr. Opin. Struct. Biol. 8: 119–223.
Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921.
Layh-Schmitt G., Podtelejnikov A., and Mann M. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146: 741–747.
Ledent P., Duez C., Vanhove M., Lejeune A., Fonze E., Charlier P., Rhazi-Filali F., Thamm I., Guillaume G., Samyn B., Devreese B., Van Beeumen J., Lamotte-Brasseur J., and Frere J.M. 1997. Unexpected influence of a C-terminal-fused His-tag on the processing of an enzyme and on the kinetic and folding parameters. FEBS Lett. 413: 194–196.
LeTilly V. and Royer C.A. 1993. Fluorescence anisotropy assays implicate protein–protein interactions in regulating trp repressor DNA binding. Biochemistry 32: 7753–7758.
Liang X.H., Volkmann M., Klein R., Herman B., and Lockett S.J. 1993. Co-localization of the tumor suppressor protein p53 and human papillomavirus E6 protein in human cervical carcinoma cell lines. Oncogene 8: 2645–2652.
Link A.J., Eng J., Schieltz D.M., Carmack E., Mize G.J., Morris D.R., Garvik B.M., and Yates J.R. III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17: 676–682.
Lopez A.J. 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32: 279–305.
Malpeli G., Folli C., and Berni R. 1996. Retinoid binding to retinol-binding protein and the interference with the interaction with transthyretin. Biochim. Biophys. Acta 1294: 48–54.
Marshall W.L., Yim C., Gustafson E., Graf T., Sage D.R., Hanify K., Williams L., Fingeroth J., and Finberg R.W. 1999. Epstein–Barr virus encodes a novel homolog of the bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J. Virol. 73: 5181–5185.
Matsudaira P. 1993. A practical guide to protein and peptide purification for microsequencing. Academic Press, San Diego, California.
Mayer A. and Barany F. 1995. Photoaffinity cross-linking of TaqI restriction endonuclease using an aryl azide linked to the phosphate backbone. Gene 153: 1–8.
Mayer M.L. and Hieter P. 2000. Protein networks—Built by association. Nat. Biotechnol. 18: 1242–1243.
Mayes A.E., Verdone L., Legrain P., and Beggs J.D. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18: 4321–4331.
McCraith S., Holtzman T., Moss B., and Fields S. 2000. Genome-wide analysis of vaccinia virus protein–protein interactions. Proc. Natl. Acad. Sci. 97: 4879–4884.
Medaglia M.V. and Fisher R.J. 2001. Analysis of interacting proteins with surface plasmon resonance spectroscopy using BIAcore. In Molecular cloning: A laboratory manual, 3rd edition (ed. J. Sambrook and D.W. Russell), pp. 18.96–18.103. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
_______. 2002. Analysis of interacting proteins with surface plasmon resonance using Biacore. In Protein–protein interactions: A molecular cloning manual (ed. E. Golemis), pp. 255–272. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Melancon P., Burgess R.R., and Record M.T. Jr. 1983. Direct evidence for the preferential binding of Escherichia coli RNA polymerase holoenzyme to the ends of deoxyribonucleic acid restriction fragments. Biochemistry 22: 5169–5176.
Mironov A.A., Fickett J.W., and Gelfand M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288–1293.
Mittler R.S., Rankin B.M., and Kiener P.A. 1991. Physical associations between CD45 and CD4 or CD8 occur as late activation events in antigen receptor-stimulated human T cells. J. Immunol. 147: 3434–3440.
Miyawaki A., Llopis J., Heim R., McCaffery J.M., Adams J.A., Ikura M., and Tsien R.Y. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388: 882–887.
Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K.N., Pfanner N., and Ryan M.T. 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8: 284–286.
Naryshkin N., Kim Y., Dong Q., and Ebright R.H. 2001. Site-specific protein-DNA photo-cross-linking. In DNA-protein interactions principles and protocols, 2nd edition (ed. T. Moss), pp. 337–361. Humana Press, Totowa, New Jersey.
Naryshkin N., Revyakin A., Kim Y., Mekler V., and Ebright R.H. 2000. Structural organization of the RNA polymerase-promoter open complex. Cell 101: 601–611.
Natsume T., Nakayama H., and Isobe T. 2001. BIA-MS-MS: Biomolecular interaction analysis for fuctional interactions. Trends Biotechnol. 19: S28–S33.
Neubauer G., Gottschalk A., Fabrizio P., Seraphin B., Luhrmann R., and Mann M. 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. 94: 385–390.
Neubauer G., King A., Rappsilber J., Calvio C., Watson M., Ajuh P., Sleeman J., Lamond A., and Mann M. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20: 46–50.
Nickel U., Chen Y.H., Schneider S., Silva M.I., Burrows H.D., and Forosinho S.J. 1994. Mechanism and kinetics of the photocatalyzed oxidation of p-phenylenediamines by peroxydisulfate in the presence of tri-2,2'-bipyridyl)ruthenium(II). J. Phys. Chem. 98: 2883–2888.
Noble D. 2002. Modeling the heart—From genes to cells to the whole organ. Science 295: 1678–1682.
Oliver S. 2000. Proteomics: Guilt-by-association goes global. Nature 403: 601–603.
Ormö M., Cubitt A.B., Kallio K., Gross L.A., Tsien R.Y., and Remington S.J. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 237: 1392–1395.
Overton M.C. and Blumer K.J. 2000. G-protein-coupled receptors function as oligomers in vivo. Curr. Biol. 10: 341–344.
Pandey A., Andersen J.S., and Mann M. 2000a. Use of mass spectrometry to study signaling pathways. Science's STKE: http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2000/37/pl1.
Pandey A., Podtelejnikov A.V., Blagoev B., Bustelo X.R., Mann M., and Lodish H.F. 2000b. Analysis of receptor signaling pathways by mass spectrometry: Identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. 97: 179–184.
Park S.-H. and Raines R.T. 1997. Green fluorescent protein as a signal for protein–protein interactions. Protein Sci. 6: 2344–2349.
_______. 2000. Green fluorescent protein chimeras to probe protein–protein interactions. Methods Enzymol. 328: 251–261.
Pearson K.M., Pannell L.K., and Fales H.M. 2002. Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun. Mass Spectrom. 16: 149–159.
Pelletier J. and Sidhu S. 2001. Mapping protein–protein interactions with combinatorial biology methods. Curr. Opin. Biotechnol. 12: 340–347.
Phizicky E.M. and Fields S. 1995. Protein-protein interactions: Methods for detection and analysis. Microbiol. Rev. 59: 94–123.
Poetsch A., Neff D., Seelert H., Schägger H., and Dencher N.A. 2000. Dye removal, catalytically active and 2D crystallization of chloroplast H+-ATP synthase purified by blue native electrophoresis. Biochim. Biophys. Acta 1466: 339–349.
Potts J.T., Young D.M., and Anfinsen C.B. 1963. Reconstitution of fully active RNase S by carboxypeptidase-degraded RNase S-peptide. J. Biol. Chem. 238: 2593–2594.
Prasher D.C., Eckenrode V.K., Ward W.W., Prendergast F.G., and Cormier M.J. 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111: 229–233.
Pugh B.F. 1996. Mechanisms of transcription complex assembly. Curr. Opin. Cell Biol. 8: 303–311.
Rain J.-C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., and LeGrain P. 2001. The protein–protein interaction map of Helicobacter pylori. Nature 409: 211–215.
Raines R.T. 1998. Ribonuclease A. Chem. Rev. 98: 1045–1065.
Raines R.T., McCormick M., Van Oosbree T.R., and Mierendorf R.C. 2000. The S Tag fusion system. Methods Enzymol. 326: 362–376. Tag fusion system. Methods Enzymol. 326: 362–376.
Rappsilber J., Siniossoglou S., Hurt E.C., and Mann M. 2000. A generic strategy to analyze the spatial organization of multiprotein complexes by cross-linking and mass spectrometry. Anal. Chem. 72: 267–275.
Richards F.M. 1955. Titration of amino groups released during the digestion of ribonuclease by subtilisin. C.R. Trav. Lab. Carlsberg, Ser. Chim. 29: 322–328.
Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., and Séraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032.
Rodi D.J. and Makowski L. 1999. Phage-display technology—Finding a needle in a vast molecular haystack. Curr. Opin Biotechnol. 10: 87–93.
Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., and Chait B.T. 2000. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651.
Ryan M.T., Müller H., and Pfanner N. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274: 20619–20627.
Sambrook J. and Russell D.W. 2001. Molecular cloning: A laboratory manual, 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Sambrook J., Fritsch E.F., and Maniatis T. 1989. Molecular cloning: A laboratory manual, 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Schägger H. 1996. Electrophoretic techniques for isolation and quantification of oxidative phosphorylation complexes from human tissues. Methods Enzymol. 264: 555–566.
Schägger H. and Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19: 1777–1783.
Schägger H. and von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231.
Schägger H., Cramer W.A., and von Jagow G. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230.
Schamel W.W. and Reth M. 2000. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity 13: 5–14.
Schreier A.A. and Baldwin R.L. 1977. Mechanism of dissociation of S-peptide from ribonuclease S. Biochemistry 16: 4203–4209.
Schuck P. 1997a. Reliable determination of binding affinity and kinetics using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 8: 498–502.
_______. 1997b. Use of surface plasmon resonance to probe the equilibrium and dynamic aspects of interactions between biological macromolecules. Annu. Rev. Biophys. Biomol. Struct. 26: 541–566.
Schwikowski B., Uetz P., and Fields S. 2000. A network of protein-protein interactions in yeast. Nat. Biotechnol. 18: 1257–1261.
Séraphin B., Puig O., Bouveret E., Rutz B., and Caspary F. 2002. Tandem affinity purification to enhance interacting protein identification. In Protein–protein interactions: A molecular cloning manual (ed. E. Golemis), pp. 313–328. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Shevchenko A., Wilm M., Vorm O., and Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858.
Sidhu S.S., Lowman H.B., Cunnningham B.C., and Wells J.A. 2000. Phage display for selection of novel binding peptides. Methods Enzymol. 328: 333–363.
Simpson R.J., Moritz R.L., Begg G.S., Rubira M.R., and Nice E.C. 1989. Micropreparative procedures for high sensitivity sequencing of peptides and proteins. Anal. Biochem. 177: 221–236.
Simpson R.J., Connolly L.M., Eddes J.S., Pereira J.J., Moritz R.L., and Reid G.E. 2000. Proteomic analysis of the human colon carcinoma cell line (LIM 1215): Development of a membrane protein database. Electrophoresis 21: 1707–1732.
Siniossoglou S., Lutzmann M., Santos-Rosa H., Leonard K., Mueller S., Aebi U., and Hurt E. 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149: 41–54.
Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., and Hurt E.C. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275.
Smith D.B. and Johnson K.S. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40.
Smith C.W. and Valcarcel J. 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25: 381–388.
Spector D.L., Goldman R.D., and Leinwand L.A. 1997. Cells: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Speicher K.D., Kolbas O., Harper S., and Speicher D.W. 2000. Systematic analysis of peptide recoveries from in-gel digestion for protein identification in proteome studies. J. Biomol. Tech. 11: 74–86.
Starr R., Willson T.A., Viney E.M., Murray L.J., Rayner J.R., Jenkins B.J., Gonda T.J., Alexander W.S., Metcalf D., Nicola N.A., and Hilton D.J. 1997. A family of cytokine-inducible inhibitors of signaling. Nature 387: 917–921.
Stryer L. 1978. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 47: 819–846.
Szöllösi J., Damjanovich S., Mulhern S.A., and Trón L. 1987. Fluorescence energy transfer and membrane potential measurements monitor dynamic properties of cell membranes: A critical review. Prog. Biophys. Mol. Biol. 49: 65–87.
Trón L., Szöllösi J., Damjanovich S., Helliwell S.H., Arndt-Jovin D.J., and Jovin T.M. 1984. Flow cytometric measurement of fluorescence resonance energy transfer on cell surfaces. Quantitative evaluation of the transfer efficiency on a cell-by-cell basis. Biophys. J. 45: 939–946.
Tsien R. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67: 509–544.
Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinisvan M., Pochart P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627.
van Roessel P. and Brand A.H. 2002. Imaging into the future: Visualizing gene expression and protein interactions with fluorescent proteins. Nat. Cell Biol. (suppl. 1) 4: E15–20.
Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351.
Verhagen A.M, Ekert P.G., Pakusch M., Silke J., Connolly L.M., Reid G.E., Moritz R.L., Simpson R.J., and Vaux D.L. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53.
Verkman A.S. 2002. Solute and macromolecular diffusion in cellular aqueous compartments. Trends Biochem. Sci. 27: 27–33.
Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., and Deshaies R.J. 2000. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11: 3425–3439.
Verveer P.J., Harpur A.G., and Bastiaens P.I.H. 2002. Imaging protein interactions by FRET microscopy. In Protein–Protein interactions (ed. E. Golemis), pp. 181–214. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Vorm O., Roepstorff P., and Mann M. 1994. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal. Chem. 66: 3281–3287.
Walhout A.J., Sordella R., Lu X., Hartley J.L., Temple G.F., Brash M.A., Thierry-Mieg N., and Vidal M. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122.
Wang L. and Dobberstein B. 1999. Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett. 457: 316–322.
Wang Y. and Stumph W. 1998. Identification and topological arrangement of Drosophila proximal sequence element PSE-binding protein subunits that contact the PSEs of U1 and U6 small nuclear RNA genes. Mol. Cell. Biol. 18: 1570–1579.
Ward W.W., Swiatek G.C., and Gonzalez D.G. 2000. Green fluorescent protein in biotechnology education. Methods Enzymol. 305: 672–680.
Weng G., Bhalla U.S., and Iyenger R. 1999. Complexity in biological signaling systems. Science 284: 92–96.
Winston F., Dollard C. and Ricupero-Hovasse S.L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55.
Wong S.S. 1991. Chemistry of protein conjugation and cross-linking. CRC Press, Boca Raton, Florida.
Wool I.G., Chan Y.-L., and Gluck A. 1995. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 73: 933–947.
Xenarios I., Rice D.W., Salwinski L., Baron M.K., Marcotte E.M., and Eisenberg D. 2000. DIP: The database of interacting proteins. Nucleic Acids Res. 28: 289–291.
Yang S.-W. and Nash H. 1994. Specific photo-cross-linking of DNA-protein complexes: Identification of contacts between integration host factor and its target DNA. Proc. Natl. Acad. Sci. 91: 12183–12187.
Young M.M., Tang N., Hempel J.C., Oshiro C.M., Taylor E.W., Kuntz I.D., Gibson B.W., and Dollinger G. 2000. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. 97: 5802–5806.
Youvan D.C., Coleman W.J., Silva C.M., Petersen J., Bylina E.J., and Yang M.M. 1997. Fluorescence imaging micro-spectrophotometer (FIMS). Bio/Technology 1: 1–16.
Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., and Cesareni G. 2002. MINT: A Molecular INTeraction database. FEBS Lett. 513: 135–140.
Zhang J.-G., Farley A., Nicholson S.E., Willson T.A., Zugaro L.M., Simpson R.J., Moritz R.L., Cary D., Richardson R., Hausmann G., Kile B.J., Kent S.B.H., Alexander W.S., Metcalf D., Hilton D.J., Nicola N.A., and Baca M. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. 96: 2071–2076.
Zimmerman S.B. and Minton A.P. 1991. Estimation of macromolecular concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 222: 599–620.
|

